AWS for Industries
Using miniwdl, GWFCore, and SageMaker Studio as a cloud IDE for genomics workflows
To keep pace with the growing scale of genomics datasets, bioinformatics scientists rely on shared analysis workflows written with portable standards such as the Workflow Description Language (WDL). The race to track SARS-CoV-2 variants notably illustrates rapid deployment of such workflows at scale on many platforms, including AWS.
This post presents a new solution for deploying WDL workflows on AWS combining miniwdl with the Genomics Workflows Core (GWFCore). miniwdl is a lightweight WDL runtime, developed by the Chan Zuckerberg Initiative, focused on productivity for command-line interface (CLI) operators and workflow developers. GWFCore is a CloudFormation template for configuring AWS Batch for large-scale genomics workloads. We developed a new AWS plugin for miniwdl to orchestrate WDL workflows on AWS Batch, with input and output data managed with Amazon Elastic File System (Amazon EFS) and Amazon Simple Storage Service (Amazon S3).
Once analysis workflows are complete, there is often a need to explore the workflow results interactively and curate them before moving to the next phase of analysis. Similarly, interactive inspection of intermediate workflow results and logs helps with debugging workflow failures.
We therefore embedded our solution within Amazon SageMaker Studio, a web-based integrated development environment (IDE) for machine learning. Adapting SageMaker Studio for WDL data processing also provides a convenient interface for interactive results curation and debugging.
Let’s see how this all fits together.
Architecture overview
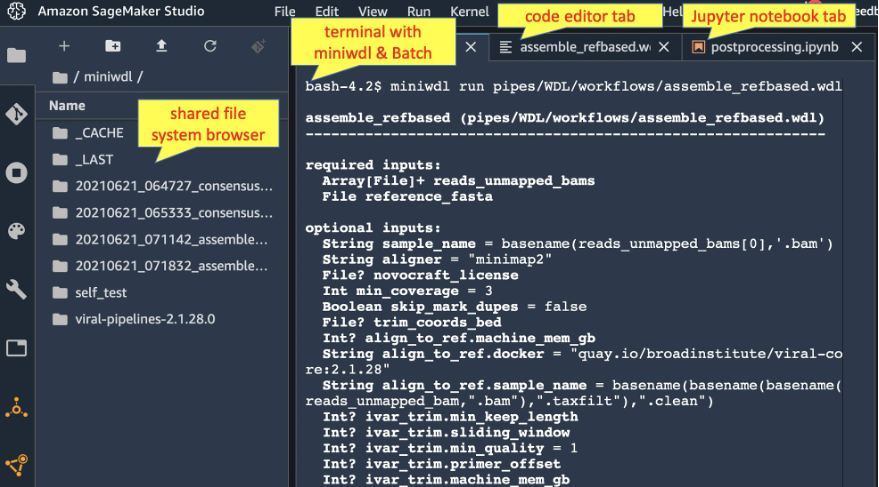
This solution uses SageMaker Studio as a web portal to launch WDL workflows on AWS Batch, manage inputs and outputs with the file system browser, and host Jupyter notebooks to explore results. The screenshot below illustrates a SageMaker Studio session with tabs for a miniwdl terminal session, WDL code editor, and Jupyter notebook, all sharing a common data view through the integrated Amazon EFS file system.
The following architecture diagram shows how miniwdl, installed in SageMaker Studio, orchestrates WDL workflows as AWS Batch tasks on a spot-instance compute environment. Amazon EFS serves as the primary file input/output location and Amazon S3 is used for long-term storage where appropriate.
Prerequisites
To try this in your own environment, you should have the following prerequisites:
- AWS account with AWS Console access
- Familiarity with WDL concepts and local miniwdl operations
Deploy the IDE
Follow these steps to deploy GWFCore integrated with SageMaker Studio, providing a command-line terminal where you can use miniwdl run to launch WDL workflows on AWS Batch.
- Create a VPC with AWS Quick Start (or use an existing VPC with public subnets)
a. Select all AZs in the desired region
b. Not needed: private subnet, NAT gateways
- Deploy a Sagemaker Studio Domain in the VPC (or use an existing domain; note that an AWS account may have only one Studio domain per region)
a. Select the SageMaker Studio button
b. Follow the “quick start” flow
i. Set a username
ii. Under execution role, select “Create a new role”
iii. In the “Create an IAM role” pop-up, select “Any S3 bucket”
c. Select the Submit button, then
i. Select your VPC
ii. Select all of its subnets
iii. Wait 5–10 minutes for the domain to deploy
d. Note your username and the Studio ID d-xxxx, shown in the SageMaker Studio Control Panel, to use as follows.
- Navigate to AWS CloudShell to deploy GWFCore using the miniwdl-aws-studio recipe
a. Run the following commands to install AWS Cloud Development Kit (AWS CDK) and the miniwdl-aws-studio recipe
Tip: You might find a newer version of miniwdl-aws-studio, with updated instructions, on GitHub.
b. Run the following command, substituting your SageMaker Studio domain ID and username:
Warning: this operation both deploys GWFCore and reconfigures SageMaker Studio domain to integrate with it. Specifically, it modifies the Studio user IAM role to allow control of AWS Batch and Amazon EFS; and the security groups to allow Studio and GWFCore to communicate. Any software you run in SageMaker Studio will inherit these permissions.
i. You will be prompted to confirm security group changes. Select yes.
ii. To customize the deployed GWFCore stack, you can add — parameters Key=Value to the cdk deploy command in order to set any of the parameters exposed by the GWFCore CloudFormation template.
- Return to the SageMaker Studio control panel, and select Open Studio.
- In the Launcher, scroll down to Utilities and files and select System Terminal to start an interactive terminal session.
- Run the following commands to install and configure miniwdl:
- Try miniwdl’s built-in self-test:
This will take several minutes as GWFCore’s AWS Batch compute environment starts up for the first time. At the end, you should see the message miniwdl run_self_test OK.
Run a viral genome assembly
Now you can use the SageMaker Studio terminal to miniwdl run WDL workflows on AWS. The ~/miniwdl directory is shared between Studio and any AWS Batch jobs submitted through a common Amazon EFS mount. With input files placed there, miniwdl run will create a run directory under which all logs and output files will appear.
For example, try the following commands in your Studio system terminal to run a viral genome assembly workflow, developed and maintained by the Broad institute, and also discussed in miniwdl’s local tutorial:
Afterwards, you can navigate the run directory under ~/miniwdl, either with terminal commands or the SageMaker Studio file system browser (look for a timestamp-prefixed directory ending in *_assemble_refbased). Inside, you’ll find the workflow.log, outputs.json referencing output files under out/, and the working directories for each WDL task invocation.
Alternatively, you can monitor the workflow operations through AWS Batch and AWS CloudWatch Logs.
Next steps
From here, you can use SageMaker Studio to orchestrate bioinformatics analyses throughout their lifecycle. You can use
- Familiar command-line tools like
aws,wget, andgitto stage input files and WDL source code miniwdlto validate workflows and launch runs on AWS Batch- Code editor to modify the WDL source code as needed
- File system browser to monitor workflow run directories and open their log files
- Jupyter notebooks to explore and postprocess WDL workflow outputs
Miniwdl can cache and reuse partial results of previous workflow runs as long as their output files remain unmodified in their run directories. But as run directories accumulate under ~/miniwdl, you may wish to use the file system browser and/or terminal commands to delete any that are no longer needed, as they incur Amazon EFS pricing.
If you plan to store output files on Amazon S3, there are two ways to transfer them. You can transfer them using aws CLI commands. Or you can automate transfer upon workflow completion using the miniwdl-run-s3upload wrapper. Try the following:
miniwdl-run-s3upload --help
Cleanup
To tear down the solution, you can destroy GWFCore’s CloudFormation stacks and delete the SageMaker Studio domain. These steps won’t remove the associated Amazon S3 bucket and Amazon EFS file system to avoid data loss. You can delete those in the respective areas of the AWS Console if needed.
Conclusion
By combining miniwdl, GWFCore, and Amazon SageMaker Studio, this solution provides familiar CLI and file system interfaces, the scalability and cost-effectiveness of AWS Batch, and convenient IDE features in Amazon SageMaker Studio, making it easy for developers and operators to build workflows and manage ad hoc analysis runs. Code for miniwdl, GWFCore, and this solution are also available as open source for those interested in tinkering with the WDL language or underlying AWS implementation details.
See our GitHub repository for the latest documentation, or open a discussion to get help.